Developer manual
Bird view
This section mainly describes how the different parts of Paroxython (code, data, results) are integrated. For details on the functioning of an individual module, click its name: it refers to the documentation page. The input and output files are linked to real examples.
Tip
On a sufficiently large screen, scroll the navigation panel until the whole program structure appears (depending on your browser, you may need to click it and use arrow keys). The following short descriptions aim to make the explanations a bit more fluid by roughly following the requirement order (imported modules and input data), from top to bottom in the diagram. Python modules appear in yellowish (extension ".py" omitted), input files and folders in reddish, and output files and databases in green.
First stage: tagging a collection of programs
source files(click to browse a copy of Python Wiki's 21 simple programs on GitHub)- It all starts with the folder you dig into when you need a Python program for your students, to use it either as an example or an exercise.
paroxython.preprocess_source- These programs are not parsed exactly as they are. The may contain textual metadata (comments, docstrings, blank lines) which are not useful for the purpose of tagging their algorithmic features, and can be suppressed. Conversely, some of their lines may entirely consist of a hint manually added by the user to denote an all-encompassing feature (e.g.,
topic:cryptography). Such manual hints need to be “centrifugated”, i.e. moved to both ends of the source, making them open on the first line and close on the last line of code. This module deals with the cleaning and the centrifugation of a given program. paroxython.list_programs- We are now able to walk through the
source filesand return the list of their preprocessed source codes. paroxython.flatten_ast- The first stage of static analysis often consists in constructing the Abstract Syntax Tree of the code to parse. Here, we go a step further by transforming this AST into a sequence of text lines, each one consisting of the whole path from the tree root to the current node. Some additional transformations are made on the fly, in order to simplify, or even make possible, the writing of regular expressions that match the algorithmic features we are interested in.
spec.md(click to browse the default specifications on GitHub)- This input file is quite special. It serves three distinct roles:
-
- As its name suggests, it specifies what features to look for, and how to find them in a given source code. The specifications are expressed either as regular expressions on the flattened version of the AST, or as SQL queries on the features found so far (either as a regular expression match or in the table resulting from a previous query).
- The Markdown format (extension
".md") is an unusual choice for a specification file. The reason for such a deviation is that this file also acts as documentation. Each formal specification is illustrated with numerous examples, and possibly with explanations, warnings, links to prerequisite or derived features, etc. - Finally, this file is also used as a test base. When
test_parse_program.pyis executed, each specification is tested on the examples provided, and its outcome checked against the expected results.
paroxython.derived_labels_db- As they are found in a given source code, the features described by
spec.mdare stored in an in-memory database, as labels with a prefix, possibly a suffix, and a span (consisting of their first and last line numbers, as well as a unique identifier to pinpoint their precise location within the AST). The storage of these labels is used dynamically to search for so-called derived features, which themselves are necessarily specified by SQL queries. paroxython.parse_program-
This module mainly consists of a function which flattens a given (preprocessed) program, parses it along the specifications defined in
spec.md, and returns the list of its labels with their spans, plus/minus those scheduled for addition/deletion.Warning. The binding of a regular expression-specified multi-line feature with its spans may reveal somewhat tricky. Make sure to understand the explanations of
get_bindings()in case your home-made regular expression yields unexpected results. paroxython.label_programs- The main function of this module not only maps
paroxython.parse_programon the list returned byparoxython.list_programs, transforming it into a list of labelled programs, but tweaks all the labels which mark an importation of an “internal” module, i.e, a program belonging to this very list. For instance, a label"import:my_program"would be transformed into"import_internally:my_program", while"import:itertools"would be left untouched. taxonomy.tsv(click to browse the default taxonomy on GitHub)-
The labels produced so far are nothing more than an intermediate result. A number of them have only been useful for deriving other labels, and can now be ignored. Those that remain must be transformed into taxa. A taxon is the structured version of one or more labels. For instance, the label
"if_without_else"will maps onto the taxon"flow/conditional/no_else", which conveniently tells us that a conditional without an else branch (no_else) is a subcase of a conditional (conditional), itself being a subcase of a control flow (flow).The file
taxonomy.tsvis a simple two-columns associative TSV array which maps label patterns onto taxa. Since some label patterns are rather long regular expressions, the columns are swapped: the second column lists the labels patterns to search; the first columns, the taxa to be substituted. For instance, the row:Taxa (replacement patterns) Labels (search patterns) flow/loop/exit/early/\11loop_with_early_exit:.+:(.+)… will trigger the following transformations into taxa:
loop_with_early_exit:for:break->flow/loop/exit/early/breakloop_with_early_exit:while:break->flow/loop/exit/early/breakloop_with_early_exit:for:return->flow/loop/exit/early/returnloop_with_early_exit:while:return->flow/loop/exit/early/returnloop_with_early_exit:for:raise->flow/loop/exit/early/raiseloop_with_early_exit:while:raise->flow/loop/exit/early/raise paroxython.map_taxonomy-
In addition to applying the transformations defined in
taxonomy.tsvto a given list of labels, the main method of this module,Taxonomy.to_taxa(), deduplicates the resulting taxa.Indeed, a same feature is often described by several labels, for instance, the
forloop of the following program:for x in range(10): print("foo") if bar(x): break -
… will be associated with no less than six labels (remember that most of them were only useful during the execution of
paroxython.derived_labels_db):"for_range:10""loop_with_early_exit:for:break""loop:for""for:x""loop_with_break:for""node:For"
The last three of these are ignored, since they do not define any mapping in
taxonomy.tsv. The remaining three are mapped respectively onto:"flow/loop/for/arithmetic""flow/loop/exit/early/break""flow/loop"
The latter is a prefix of (at least) another one: as an overly broad categorization of the feature, it can be discarded. Only the former two will go into the deduplicated result: they indeed categorize the feature along two partially distinct dimensions (the iteration domain and the nature of the exit).
paroxython.make_db- This is the main user-facing module of the first stage, invoked on command line by:
paroxython collect DIRECTORY -
It walks through the input directory, labels its programs and maps these labels onto the default taxonomy (or yours). Along this process, the labels
"import_internally", calculated byparoxython.label_programs, are used to list, for each program, those of the input directory that it imports or by which it is imported (either directly or indirectly).Finally, the results are stored as a NoSQL (JSON) or relational (SQLite) database.
*_db.json(click to browse the tag database of 21 simple programs).-
Generated by
TagDatabase.get_json()as a dictionary (or, in JSON parlance, object) with the following keys:"programs": maps each program path to the timestamp of its last modification, its cleaned up source code, the list of its labels with their poor2 spans, and that of its taxa with their poor spans;"labels": maps each label to the programs that feature it;"taxa": maps each taxon to the programs that feature it;"importations": maps each program to the programs that import it;"exportations": maps each program to the programs by which it is imported.
Note that the labels are not used thereafter: they are stored only for logging purposes.
*_db.sqlite- An experimental relational version of the above JSON tag database. Also generated by
paroxython.make_db, but currently unused. SeeTagDatabase.write_sqlite()for its schema and an example query. Not under version control.
Second stage: getting recommendations
*_pipe.py(click to browse an example pipeline to be executed on the database of 21 simple programs)- To get program recommendations, you first create a pipeline, i.e., a Python file consisting in a list of filtering commands to be applied on a JSON tag database generated by
paroxython.make_db. Each command is a dictionary with two keys: -
"operation": a string among"include","exclude","impart"and"hide", with an optional suffix" any"(implicit) or" all"."data": either a string consisting in a shell command, or an heterogeneous list of criteria: patterns (matching either program paths or taxon names) or semantic triples (of the form subject, predicate, object).
-
The interpretation of a predicate is not immediate, and is the object of the next two modules.
paroxython.compare_spans- A dictionary of predicates listing all possible relations between two intervals (x_1, x_2) and (y_1, y_2). The keys of these predicates are expected to be used as the second term of a semantic triple expressing a relationship between the spans of two features. Generated by
make_compare_spans.py. paroxython.normalize_predicate- A thin layer between
paroxython.compare_spansandparoxython.filter_programs, this function tries to recognize as a key a predicate given by the user. In the case the predicate is negative (e.g.,"not inside"), strip it from the predicate ("inside"). Returns the corresponding lambda function along with a boolean indicating whether it was negated or not. paroxython.filter_programs-
Probably the most complex part of Paroxython. A filter instance is initialized with a JSON tag database of tagged programs. It is characterized by the state of the following attributes:
"selected_programs": the programs to be recommended, initially all of them."imparted_knowledge": the concepts (i.e., taxa) that have already been presented to your students, and therefore have a learning cost of zero. Initially empty."hidden_taxa"/"hidden_programs": the programs and/or taxa you are not interested in seeing in the resulting tables. It's just a display thing, with no effect on the actual filtering. Initially empty.
The filter responds to the pipeline commands sent to
ProgramFilter.update_filter()by evolving the contents of these three sets. paroxython.assess_costs- The costs in question are the learning costs. The final state of the imparted knowledge calculated by
paroxython.filter_programsis used to initialize the constructor after all the pipeline commands have been executed. The main methodLearningCostAssessor.__call__()is then invoked with the remaining recommended programs. paroxython.recommend_programs- The user-facing module of the second stage, called on command line by:
paroxython recommend -p pipeline_path DB_PATH -
A simple wrapper around
ProgramFilter, with report capabilities. For each command of the pipeline, it checks the validity of its operation and data, and sends them toProgramFilter.update_filter(). When the pipeline is exhausted, it invokesLearningCostAssessor.__call__()to compute the relevant learning costs. -
At the end, a call to
Recommendations.get_markdown()creates a report of the results. *_recommendations.md(click to browse the report of the execution of the above pipeline)- The final report sorts the recommended programs by increasing learning cost and SLOC, and group them into predefined cost intervals3. It includes a table of contents, and a summary of which program has been filtered at what stage of the pipeline execution.
Helper programs
During the development process of Paroxython, it proved useful to create a number of additional scripts to make the final program easier to write, test or understand. None of these scripts are meant to be run by the end user: as such they are are not included by pip install. But if you have to modify the source code to better meet your personal expectations, or are interested in collaborating on the project, some of them are definitely a must4.
Code generators
make_compare_spans.py- Generates the source code of the module
paroxython.compare_spans. Must be launched manually. reformat_spec.py- Reads the contents of
spec.md, normalizes its formatting (Markdown and SQL) and writes it back. The goal is to ensure the consistency of the document and the validity of its numerous internal links, as well as to avoid authors wasting time with formatting details. Launched automatically bytest_parse_program.py. suggest_regex.py- The regular expression patterns of
spec.mduse a rather limited and repetitive array of techniques. This program aims to partially automate their production. Given theflat_ast.txtoutputted byparse_source.py(cf. testing helper below), extract the lines that characterize the feature to be recognized, and save them in thesandboxfolder under the nameflat_ast_selection.txt. The script will then print a regular expression that matches this selection.
Testing
dump_flat_asts.py- Extracts from
spec.mdall the features specified by a regular expression, useflatten_ast()to parse their examples into a flat AST, and outputs the results assandbox/flat_ast.txt. make_dummy_db.py- Generates
taxa_and_programs.txtand, from this file,programs_db.json, used as a dummy example in various tests. make_programming_idioms_folder.py- Populates the example directory
idioms/programs, with the broad variety of code snippets snapshotted from the amazing Programming Idioms website of Valentin Deleplace. More infohere. parse_source.py- A wrapper around
paroxython.parse_programthat allows you to quickly test the labelling of a given source code. This one must be placed under the namesource.pyin asandboxdirectory (unversioned). The flattened version of its AST is generated asflat_ast.txtin the same folder, while its label table is displayed in Markdown format. All of this may streamline the work onspec.md.
Documentation builders and examples
build_pdoc.py- Builds this documentation with the excellent pdoc3 command line tool, and applies a number of patches to tailor the result to our needs.
draw_flow.py- Generates the diagrams on the present page and
paroxython.filter_programs. Launched automatically bybuild_pdoc.py. example.sql- An example of query on the experimental SQLite database generated by
paroxython.make_db. parse_syllabus.py- This script is invoked in the first example of the pipeline documentation. This is the one that the main author of Paroxython relies on to automate the extraction of the “imparted” programs from a textual timeline he updates after each lesson. The script starts by dropping all the text following the
"# EOFtag. In the remaining part, it matches all the Python program paths which are preceded by a"+"symbol and possibly a bracketed string (for instance"+ [mandatory] homework.py"). Of course, if you use other conventions, you will have to come up with your own custom script. It can be written in any language (e.g., Perl, bash, etc.), as long as it prints a list of program paths on stdout. update_tree.py- Computes a taxonomy of The Algorithms - Python (not versioned here) and generates
tree.js, a JavaScript list ready to send to the Google Chart API used to draw this word-tree. Launchbuild_pdoc.pyafterwards.
Tag databases
Depending on the extension of DB_NAME (either ".json" or ".sqlite"), the command:
paroxython collect -o DB_NAME DIRECTORY
… will create two kinds of databases, each with their own purpose.
The JSON tag database
It is this type of so-called NoSQL database that Paroxython draws upon to recommend programs. Its schema is:
{
"programs": {
"program_1.py" : {
"timestamp": "1970-01-01",
"source": "print('hello')\nprint('world')\n",
"labels": {
"label_1": [[span_start_1, span_end_1], ...],
...
},
"taxa: {
"taxon_1": [[span_start_1, span_end_1], ...],
...
}
},
...
}
"labels": {
"label_1": ["program_1.py", "program_2.py", ...],
...
},
"taxa": {
"taxon_1": ["program_1.py", "program_2.py", ...],
...
},
"importations": {
"program_1.py": ["program_2.py", "program_3.py", ...],
...
},
"exportations": {
"program_1.py": [],
"program_2.py": ["program_1.py", ...],
...
}
}
As an example, take a look on the JSON tag database of the Python Wiki's 21 simple programs.
The SQLite tag database
This kind of database is fundamentally agnostic, but lends itself quite well to statistics. Its schema is:
CREATE TABLE program (
program TEXT PRIMARY KEY,
timestamp TEXT,
source TEXT
);
CREATE TABLE label (
-- use rowid as primary key
label TEXT,
label_prefix TEXT,
label_suffix TEXT,
span TEXT,
span_start INTEGER,
span_end INTEGER,
program TEXT,
FOREIGN KEY (program) REFERENCES program (program)
);
CREATE TABLE taxon (
-- use rowid as primary key
taxon TEXT,
span TEXT,
span_start INTEGER,
span_end INTEGER,
program TEXT,
FOREIGN KEY (program) REFERENCES program (program)
);
Having a relational version of the tag database means that it can be queried with SQL. Here is an example which returns the 20 most frequent taxa:
SELECT taxon, count(*) AS occurrences
FROM taxon
GROUP BY taxon
ORDER BY occurrences DESC
LIMIT 20
More complex, a query comparing the number of occurrences of the loops for and while:
SELECT (CASE SUBSTR(taxon, 11, 3) -- extract either "for" or "whi"
WHEN "for" THEN "for" -- and replace it either by... itself
ELSE "while" -- or by the complete name of the loop
END) AS loop,
COUNT(*) AS occurrences
FROM taxon
WHERE taxon LIKE "flow/loop/for%"
OR taxon LIKE "flow/loop/while%"
GROUP BY SUBSTR(taxon, 0, 14) -- cut the taxon three characters after "flow/loop/"
In theory, the relational database can also be used to select programs according to certain criteria:
SELECT
taxon,
program,
group_concat(span, ", ") AS spans,
source
FROM program
JOIN taxon USING (program)
WHERE taxon GLOB "type/non_sequence/dictionary/*"
GROUP BY taxon, program
On the provided simple programs, the execution yields:
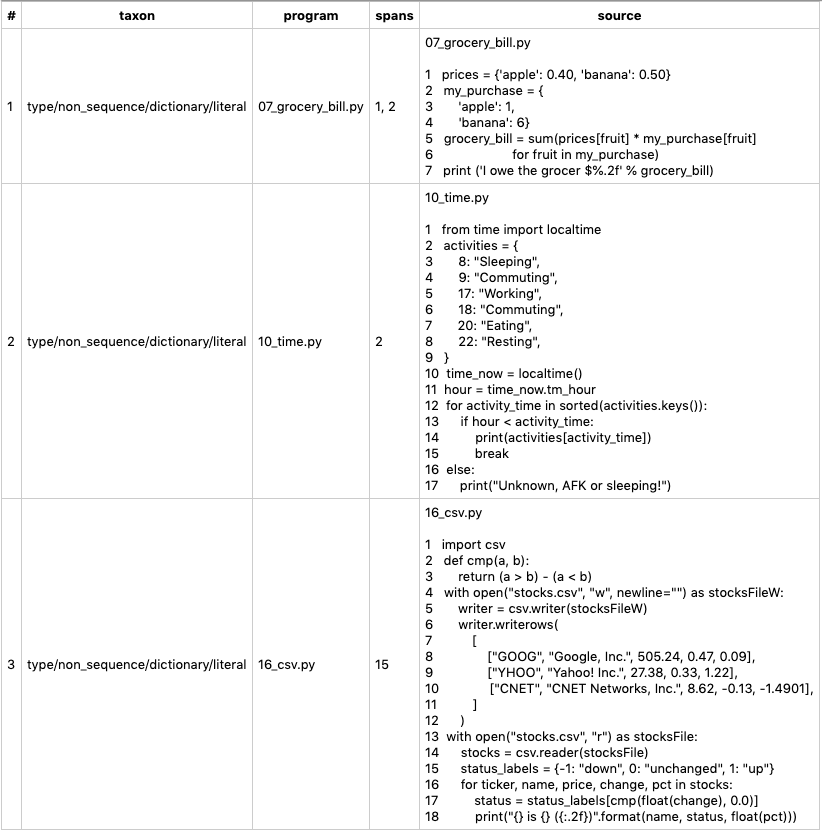
Note however that the same list of programs (with the nice addition of learning costs) could have been obtained by feeding the function Recommendations.run_pipeline() with an elementary command:
{
"operation": "include",
"data": ["type/non_sequence/dictionary/"]
}
So, although SQL should be familiar to almost everyone in our target audience, and might “make complex things possible”, as far as filtering is concerned, our minimalistic schema arguably does not ”make simple things simple” (to paraphrase Alan Kay). Some denormalization should ease the process, but so far we have prioritized the development of the pipeline system.
Warning
Currently, the relational database is better suited for making statistics than recommendations.
Implementation notes
User types
Paroxython makes heavy use of Python 3.5+ type hints. They are documented directly in user_types.py.
Default argument trick
Several functions or methods declare a compiled and bound regex pattern as an optional argument (see normalize_predicate() for an example). Such arguments are not meant to be provided by the caller. Their default value will be used systematically, with the benefit of being evaluated only once. This is arguably better for both performance and encapsulation. More details on Stack Overflow.
-
In the replacement pattern,
"\1"denotes the 1st captured group (in parentheses) of the search pattern. ↩ -
The poor span of a feature's occurrence is its span, deprived from the last member (location in the AST). ↩
-
The intervals are predefined as [0, 0], ]0, 0.25[, [2^{i-2}, 2^{i-1}[, \dots for all natural numbers i. ↩
-
However, in the current state of things, please don't expect the
Spanish inquisitionsame level of code quality, documentation and testing than in the user-facing parts of the package. ↩